Design approach
The Artiatomi package was designed around a core algorithmic part that entirely runs on GPU making use of Nvidia’s CUDA framework. As especially operations on larger volume datasets have a huge memory consumption, many tasks can be split over multiple GPUs using MPI. This allows on the one side the parallel execution on multiple compute nodes (one single multi-GPU computer or even an entire network as a compute cluster) to speed-up computation. But also allows for large memory allocations to process large volume datasets without physical memory limitations.
Artiatomi comes thus as two different kinds of tools: a simple command line-based interface for most applications in the package and a graphical user interface for aiClicker and aiCtfDetectorGui. aiClicker further doesn’t make use of any CUDA-acceleration and can therefore be used on any computer, from a simple laptop to a powerful workstation. All command line interfaces are controlled using configuration files, i.e., simple text files that contain all parameters needed for a specific processing step.
In general, the computer configuration is as follows: One or more power-full GPUs are available in one or more computers as a GPU compute cluster and all computers can access a common file server. The user connects to one of these compute nodes via SSH and runs a process. The input data is configured and the computed result is then inspected on a standard PC independently of the GPU compute nodes.
Metadata
No application of the Artiatomi package modifies any of the existing files of a dataset: All files are accessed as read only. In order to store necessary metadata for processing, Artiatomi creates an appendix file with the same name plus “.info” as additional extension. For each kind of image file (movie stack, tilt series, volume, etc.), the data contained in such an info file varies. The file itself is a simple JSON file that can be read and written by basically any software, such as MATLAB, Python-scripts, and many more.
For movie stacks, Artiatomi stores the shift information for individual frames in its metadata file. A tilt series would contain all its alignment information, dose and CTF-parameters. Volumes store information about the reconstruction process, additional shifts and tilts, and parameters for the missing wedge.
If no .info file is available, the image file header is used to extract as much information as possible to create a new metadata file. But for correct parameters, a full data import is necessary in order to obtain best possible results.
The only exception from this scheme are motivelists and shift files:
Motivelists
Motivelists are files that store for one or more tomograms the position and orientation for sub-volumes. The motivelist is used to visualize a protein of interest inside the entire tomogram, or to average many sub-volumes with the correct orientation in order to obtain a high-resolution sub-tomogram-average.
Motivelists can be stored in three different formats, whereas the filename-extension determines the format to use:
- .motl — a binary format used inside Artiatomi. The fact that the data is stored binary reduces the file size in case of large datasets.
- .json — the same format as .motl and allows the same features as .motl, but the file is stored in text-JSON-format. This can be useful for manual editing and testing, but too slow for large datasets.
- .em — this is the compatibility format for legacy tools based on the EM-Toolbox. Not all features are supported in this format, for example the post- and pre-reconstruction displacements are always merged together in this format. Use this format only for data im- or export to other packages but not for processing inside Artiatomi.
Artiatomi uses global coordinates to position individual sub-volumes in space, independent of any shifts or binning. Legacy and other tools based on the EM-Toolbox use a voxel-based coordinate instead. Use the aiToolbox to convert a motivelist from one coordinate system to another. For example, to import a motivelist and to convert from voxel coordinates to Artiatomi global coordinates:
aiToolbox motivelist tomoToGlobal -i motivelistToImport.em -o motivelistInArtiatomi.motl -t the_reconstructed_tomogram.emShiftfiles
Shiftfiles are binary files that contain the local shift on each micrograph/tilt for each sub-volume of a tomogram. These files are created during shift refinement and used during reconstruction based on the refinement. As a shiftfile is not meant to be edited by the used manually, the file is binary.
Configuration files
As mentioned before, the command line-based tools operate with configuration files to pass the algorithmic settings to the application. A configuration file is a text file where each line defines a parameter in the form “parameterName = value”. The application checks the parameters for consistency and for correct value ranges (if applicable) and also replaces numeric place holders and expands relative path names.
Some parameters can also be provided by the command line, too: In the usual UNIX/LINUX form --parameterLongName value or -parameterShortName value. Note that if parameters are passed by configuration file and by command line, the command line overrules the file parameter. To obtain a list of possible command line parameters, each application can be executed with the --help or -h argument. In order to obtain a sample configuration file with all possible parameters, execute the application with the --example filename.cfg or -ex filename.cfg argument and the parameters with their default values are stored in a file named “filename.cfg”
The combination of numeric place holders and command line arguments allows for having at the same time a common set of processing parameters for large datasets and specific adaptions for individual files. Further, if filenames are given as a relative path (first character is not ‘/’ on Linux), all filenames passed by configuration files are relative to the input file given by the “Input” parameter.
As an example, we can define one reconstruction configuration file for an entire dataset of multiple tilt-series in the following way:
…
Input =
Output = rec_##_SART.em
ReconstructionMethod = SART
OverSampling = 2
…If we call the reconstruction given this configuration and pass the input file – the tilt series to reconstruct – via command line using the -i or --input argument, we will obtain in the folder of each input file a reconstruction file ‘rec_##_SART.em’ where ‘##’ will be replaced by the tomogram number.
If we would set
Output = ../SART_reconstructions/rec_##.emand all tomograms are given in a directory structure as described below, we would obtain all reconstructions in a common folder ‘SART_reconstructions’.
Numeric placeholders are either given by ‘##’ or ‘#2#’, where the number in-between the ‘#’ determines the length of the resulting character chain which is filled with leading zeros in case it is shorter. For example: a tomogram number of ‘12’ and a filename given as ‘filename_#4#.em’ would be turned into ‘filename_0012.em’. These placeholders can be provided at any place in the path and file name.
Further, some files have a specific naming scheme, for example references in sub-tomogram averaging are always named ‘something_referenceNr_iterationNr.em’ or ‘.mrc’. For these naming-scheme numbers, any number or the ‘#’-symbol are replaced by the actual number as given from the current processing context. The configuration file thus doesn’t need to be adjusted for each iteration in the given example.
Directory structure
To make best use of the configuration file system, the following directory structure is recommended:
…/someDirectoryToTheProject/
tomograms
tomo_1
tomo_2
tomo_3
…
moviestacks (movie stacks can also be stored in the same directory or a sub-directory as tomograms)
stack_1
stack_2
stack_3
averaging
masks
motivelists
particles
references
Global coordinate system
Except for algorithms used for dose fractionation stack alignment where only a two-dimensional relative shift is determined, Artiatomi always operates in a three-dimensional coordinate system which is the same throughout the entire processing – independent of any alignments or data binning: The reference is always the un-shifted, un-tilted and un-binned coordinate system defined as in
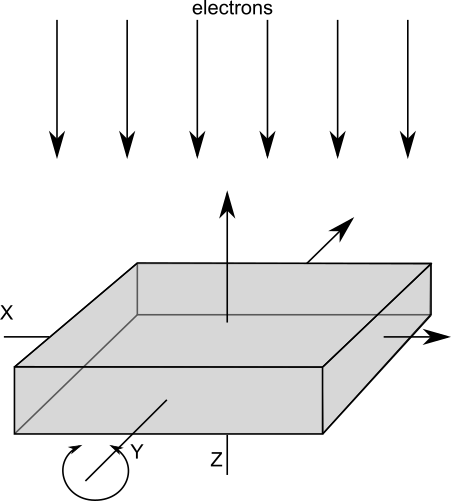
If a reconstruction is performed on a binned or un-binned tilt-series doesn’t matter: they both use the same alignment parameters and result in the same volume geometry for the reconstruction. Also, the position of sub-tomograms is internally stored as the absolute position in that global coordinate system and not for example relative to some voxel coordinates. Further, a motive list — a specific file that stores position and orientation of individual sub-volumes — does not depend on particle binning either: Shifts and positions are always given in this global reference system.
Fourier filtering
Basically, all processing is done on Fourier-filtered data and filter parameters must be provided: low-pass and high-pass limit and a sigma range to smooth the filter edge. Depending on each algorithm, either a Gaussian-decay is used or a cosine-shaped drop-off is used. The filter-parameters can be passed in two different ways: as relative values or as absolute values. If relative values are used, they indicate the fractional part of the spectrum to use ranging from [0..0.5] with 0.5 being the Nyquist frequency (of the unbinned image). Absolute values are given in pixels and define the pixel-distance in the power spectrum and range from [0..imageDimension/2]. Thus, if all filter parameters are given in the range [0..1[ they are taken as relative values, otherwise as absolute values and are converted to relative ones based on the un-binned image dimension (if not specified differently). For non-square images, the larger dimension is used.
Supported file types
Artiatomi supports most commonly used filetypes used in the field of cryo-EM for input images. MRC-typed images, stacks and volumes are supported, also the EM file format is commonly used. Further TIFF files can be read and written to, with or without compression. Also, the DM3 and DM4 file format is supported for reading, such as the SER format.
Input files can have nearly any pixel datatype, for processing all images are converted to float internally. Some applications let the user choose the pixel format for images to save whereas volumes are always stored as float.
Support for MPI
Many applications in Artiatomi support the use of multiple GPUs on multiple hosts in parallel. To leverage the power of multiple GPUs, simply run the corresponding application using mpiexec, for example:
mpiexec -n NumberOfNodes aiRec -u settings.cfgMagnification Anisotropy
Some other cryo-electron microscopy packages, especially for single particle, do not directly compensate for magnification anisotropy. Instead this is another optimization variable among others and is thus only taken into account while CTF/defocus determination.
In Artiatomi this approach is not possible: magnification anisotropy must be known before any processing and is a user input. Artiatomi compensates magnification anisotropy already during tilt-series alignment, which for example allows optimizing for beam-declination within the marker model. But the main reason for this approach is 3D-CTF-correction: As Artiatomi corrects for the CTF in the entire volume with a varying defocus depending on the Z-distance, CTF astigmatism must be distinguished from magnification anisotropy. Otherwise the thon-rings would run out of phase when applying a defocus offset.